|
|
|
| Home » Transcription » Research |
| Transcription |
Current Research Interests
The transcription laboratory is engaged in understanding the mechanism, physiology, and inhibition of bacterial Rho-dependent termination. A wide range of techniques from biophysics (spectroscopy, thermodynamics, fast kinetics, etc.), biochemistry (protein purification, chemical and enzymatic foot-printing of protein and nucleic acids, cross-linking, etc.), molecular biology (recombinant DNA techniques, site-directed mutagenesis, super-resolution microscopy), bacterial genetics and genomics are used in the laboratory to solve these intellectually challenging problems.
Projects:
- Mechanism of transcription termination by transcription termination factor Rho.
- Mechanism of Rho-NusG interaction in vivo and in vitro.
- Physiological roles of Rho-dependent terminations.
- Super-resolution microscopy of the transcription machinery.
- Isolation of mycobacteriophage-derived proteins with antimicrobial activities.
- Design of antimicrobial peptides from bacteriophage proteins
Collaborators::
-
Prof. Markus Wahl, Freie Universität Berlin, Germany (Cryo-EM).
-
Prof. Udaydittya Sen, SINP, Kolkata (Crystallography).
-
Prof. Agnieszka Szalewska-Pałas, Uniwersytet Gdański, Poland.
-
Prof. Jayanta Mukhopadhyay.
Research Highlights |
|
The Psu protein of phage satellite P4 inhibits transcription termination factor ρ by forced hyper-oligomerization (Nature Communication 2025; in collaboration with Markus Wahl).
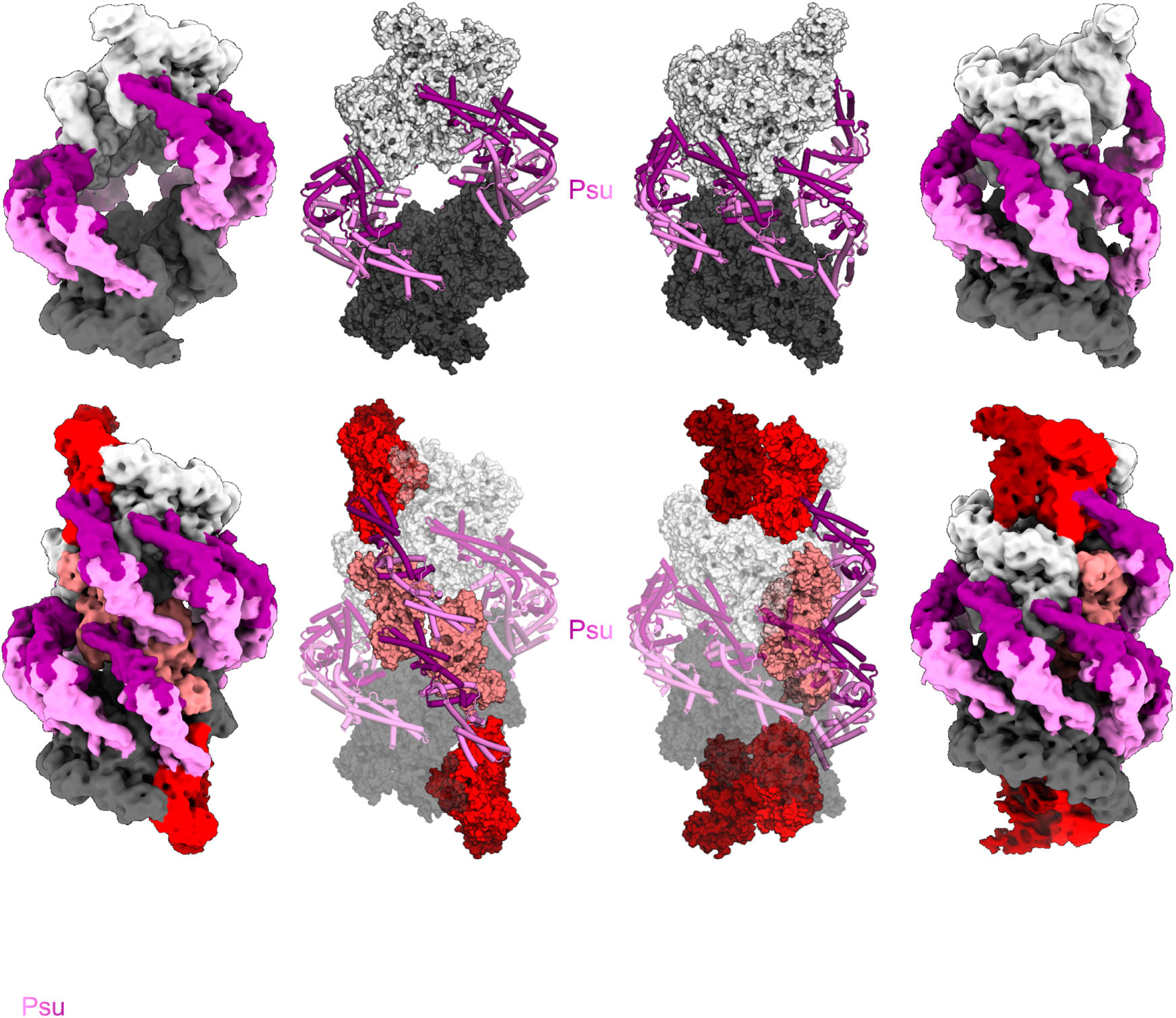
Many bacteriophages modulate host transcription to favor the expression of their genomes. Phage satellite P4 polarity suppression protein, Psu, a building
block of the viral capsid, inhibits hexameric transcription termination factor, ρ, by presently unknown mechanisms. Our cryogenic electron microscopy structures of ρ-Psu complexes show that Psu dimers clamp two inactive, open ρ rings and promote their expansion to higher-oligomeric states. ATPase, nucleotide binding, and nucleic acid binding studies revealed that Psu hinders ρ ring closure and traps nucleotides in their binding pockets on ρ. Structure-guided mutagenesis in combination with growth, pull-down, and termination assays further delineated the functional ρ-Psu interfaces in vivo. Bioinformatic analyses revealed that Psu is associated with a wide variety of phage defense systems across Enterobacteriaceae, suggesting that Psu may regulate the expression of anti-phage genes. Our findings show that modulation of the ρ oligomeric state via diverse strategies is a pervasive gene regulatory principle in bacteria.
|
|
Functionally important components of the transcription elongation complex involved in Rho-dependent termination. (FEMS Microbiology Letter, 2025). 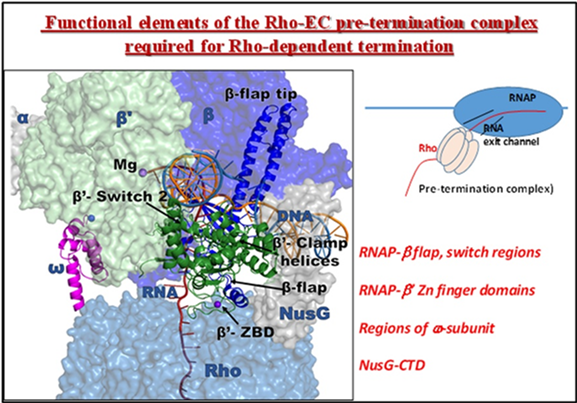
Bacterial transcription terminator, Rho is an RNA-dependent ATPase that terminates transcription. Several structures of pre-termination complexes of the Rho-transcription elongation complex (EC) revealed a static picture of components of the EC that come close to the nascent RNA-bound Rho, where many of the residues of EC reside ≤10Å from the Rho residues. However, the in vitro-formed Rho-EC complexes do not reveal the dynamic interaction patterns of the in vivo Rho-EC during the termination process. Here we report synthetic defect analyses of various combinations of the mutations in RNAP ’ and -subunits, NusA, NusG, and Rho proteins to delineate the functional network of this process. Several mutations in the -flap and ’-Zn-finger and -Clamp helices domains of RNAP are synthetically defective in the presence of Rho mutants indicating functional involvement of these domains. Mutations in the NusA RNA-binding domains were synthetically defective with the Rho mutants suggesting its involvement. Our genetic analyses also revealed functional antagonisms between the -subunit of RNAP and the NusG-CTD during termination. We concluded that the regions surrounding the RNA exit channel, the RNA-binding domains of NusA, the RNAP -subunit, and NusG-CTD constitute a functional network with Rho just before the onset of in vivo Rho-dependent termination.
.
|
|
DNA binding of an RNA helicase bacterial transcription terminator (Biochemical Journal, 2025).
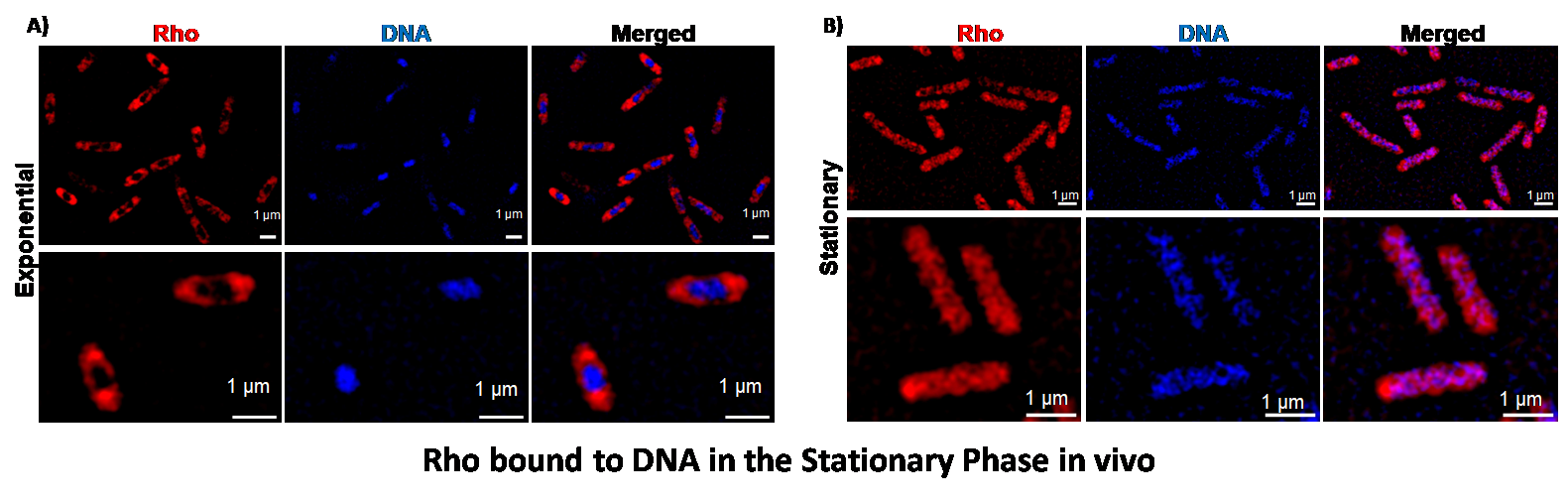
The bacterial transcription terminator Rho is a hexameric ATP-dependent RNA helicase that dislodges elongating RNA polymerases. It has an N-terminal primary RNA binding site (PBS) on each subunit and a C-terminal secondary RNA binding site at the central channel. Here, we show that Rho also binds to linear longer double-stranded DNAs (dsDNA) and the circular plasmids non-specifically using its PBS. However, this interaction could be competed efficiently by single-stranded DNA, dC34. Long dsDNA (3.5 kb) at the PBS activates short oligoC RNA-mediated ATPase activity at the secondary binding site (SBS). The pre-bound Rho to this long DNA reduces the rate and efficiency of its transcription termination activities in vitro. Elevated concentrations of Rho reduced the in vivo transcription level suggesting that Rho might also function as a non-specific repressor of gene expression under certain conditions. In the mid-log phase culture, Rho molecules were concentrated at the poles and along the membrane. In contrast, the Rho hexamers were observed to be distributed over the bacterial chromosome in the stationary phase likely in a hyper-oligomeric state composed of oligomers of hexamers. We propose that Rho molecules not engaged in the transcription termination process could use the bacterial chromosome as a “resting surface”. This way the “idle” DNA-bound Rho molecules could be kept away from accidentally loading onto the nascent RNA and initiating unwanted transcription termination.
|
|
Projects in progress:
-
Understanding the physiological consequences of Rho-dependent termination.
-
Understanding the role of the omega subunit of RNAP in Rho-dependent termination.
-
Super-resolution Microscopy of Rho.
-
Isolation and characterization of anti-mycobacterial proteins from mycobacteriophages.
-
Design of peptides from bacteriophage proteins.
Extramural Funding:
- SERB JC Fellowship (2022-2027)
- SERB grant (2024-2027)
- DBT Grant (2022-2025)
Awards/Recognition:
- 2002-2007: GRIP research grant award from NIH, USA.
- 2003-2008: Wellcome Trust, UK, Senior Research Fellowship.
- 2007: DBT Bioscience carrier development award.
- 2007: Elected member of GRC.
- 2008: DST Swarnajayanti Research Fellowship.
- 2011: Elected fellow of NASI, Allahabad.
- 2015: Member DST- SERB, task force.
- 2018: Elected Fellow INSA, New Delhi.
- 2018: Elected fellow IASc, Bangalore.
- 2018: Fellow of Telangana Academy of Sciences.
- 2019: DBT TATA Innovation Fellowship.
- 2021: Elected Fellow of West Bengal Academy of Sciences.
- 2022: SERB JC Bose Fellowship.
Editorial Board Member:
- Journal of Applied Genetics (microbial Genetics section).
Reviewer of Journals/grants/Thesis:
Nature Communications, Cell Genomics, MBio, TIBS, Journal of Bacteriology, Journal of Molecular Biology, Microbiology, PLOS one, Biochemical transactions, Communication Biology, msphere, Transcription MEEGID, Microbial Genomics, Scientific Reports, Indian Journal of Biophysics and Biochemistry, Journal of
Bioscience, etc.
The reviewer of grants for different granting agencies like DBT, DST, DRDO, Indo-French programs, Marsden Fund, New Zealand. etc.
The reviewer of the thesis of the Ph.D. students from renowned institutions like SINP, Kolkata; Bose Institute, Kolkata; IISc., Bangalore; IMTech, Chandigarh, HCU, JNU, etc.
Patents :
- 'NOVEL SYNTHETIC PEPTIDES'; Indian Patent Application No. 201841048582 filed on December 20, 2019.
| |
|
|
|
| Last updated on : Friday, 29th April, 2022. |
|
|
|
|